Abstract
Background and Aim: Multiple Myeloma development is under polygenic control and as an evidence for inheritance, we have recently reported positive and negative HLA-myeloma associations with: HLA-C*07:02g (OR:1.27, p=5.27 x 10 -7) and HLA-C*05:01 (OR: 0.85, p=0.039)(Beksac et al Leukemia 2016:1-4). HLA-C*07:02g (Group C1) and HLA-C*05:01 (Group C2) are listed among the KIR ligands, These ligands usually interact with inhibitory KIRs(iKIR) while having weaker binding with activating KIRs(aKIR). KIRs play a major role in regulation of immunity and self-tolerance. Attempts to inhibit the inhibition exerted by immune check point regulators is an evolving field. Such monoclonal antibodies in combination with agents augmenting NK cell activation have exerted acceptable responses. However the distribution of KIRs or their ligands have not been studied extensively in myeloma. The aim of this study was to analyze the frequency of KIR genotypes and their cognate ligands among myeloma patients comparing with healthy controls.
Patients, controls and treatment: 117 myeloma patients admitted to our clinic and consented between Sept 2016-July 2017 were enrolled in the study. Patient characteristics were: 34- 91(median: 61), ISS:I/II/III:46 /45 /25 ; IgG/IgA/Light chain: 67/18 /20 . 65 % of patients had received at least one ASCT. 68 of myeloma cases were on long term remission of min 2-8 years. 49 patients had experienced either early progression within 60 days from the induction or multiple relapses. As a control group 215 healthy subjects and related/unrelated hematopoietic stem cell donors aged 18-65 and screened for HLA and KIR genotyping were included. All controls had KIR and HLA typing but only 172 had HLA-C data available for Group C1 and C2 assessments.
KIR and KIR ligand genotyping: The Olerup SSP KIR Genotyping Kit (Olerup, Stockholm, Sweden) enabled us to detect 16 KIRs: KIR2DL1, 2DL2, 2DL3, 2DL4, 2DL5A/B, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, 3DL2, 3DL3, 3DS1, 2DP1, 3DP1. The Olerup KIR HLA Ligand Detection Kit was used to type HLA-ABw4+, HLA-BBw4+Thr80, HLA-BBw4+Ile80, HLA-BBw4+Asp77,Thr80, HLA-CAsn80, and HLA-CLys80.
Statistical analysis: The Statistical Package for the Social Sciences (SPSS 11.5 for Windows, IBM Statistics, USA) was used for data analysis. Pearson's chi-square test and Fisher's exact test were applied for categorical variables. For continuous variables, t-test was applied. Variables found positive in all subjects of both groups were excluded from analysis.
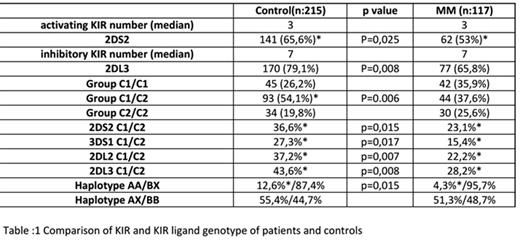
Results: Comparison of healthy controls and all myeloma subjects are summarized in Table1. Myeloma patients had similar median number of aKIR and iKIRs. There were more iKIR (n:7) genes expressed than aKIRs (n:3) among normals and patients. However there were less cases expressing haplotype AA among the myeloma group compared to healthy controls( 4.3 vs 12.6%). Overall iKIRs were expressed widely among controls ( 66-100%) and myeloma (57.2-100%) cases. Among the iKIRs only 2DL3 was found to have a statistically significant difference : less frequent among patients. As for aKIRs only 2DS2 was significantly different: less frequent in myeloma. The ligand for both 2DL3 and 2DS2 is HLA-Group C1. We were able to detect Group C1/C2 heterozygosity to be less frequent and a trend to increase in the frequency of C1 or C2 homozygotes among myeloma patients. Two aKIRs 2DS2, 3DS1 and two iKIRs 2DL2,2DL3 were less frequent among C1/C2 heterozygous myeloma patients.
Conclusion: This study, to our knowledge, is the first to compare KIR and KIR ligand genotype frequencies between myeloma patients and healthy controls. The significant difference between patients and controls was a less frequent observation of an aKIR and/or iKIR among patients. The trend to increase in HLA group C1 homozygosity, is compatible with our previous HLA-Myeloma association study. In this preliminary analysis, from one geographical region, we were not able to detect a strong correlation between KIR genotype and myeloma. In future we are planning to analyze the outcome of patients with distinct iKIR or aKIR profiles.
Acknowledgment: The study has been supported by Scientific and Research Council of Turkey (TUBITAK grant no: 115S579) and Turkish Academy of Sciences.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal